
SMAG has advanced its internal drug repurposing research efforts with the development of version 2.1 of its integrated knowledge graph and computational analysis framework. This internally built system supports SMAG’s scientific teams in identifying potential new therapeutic applications for existing drugs. By combining structured biomedical data, relationship modeling, and domain expertise, SMAG’s drug repurposing initiative offers a scalable and rigorous approach to uncovering novel treatment opportunities.
The Value of Drug Repurposing
Drug repurposing — the process of identifying new medical uses for already approved or investigational drugs — has become a valuable strategy for accelerating the development of therapies. Because repurposed drugs have already passed many safety and toxicity evaluations, they may reach clinical trials and regulatory approval more quickly than novel compounds.
Despite its potential, identifying repurposing opportunities requires navigating vast and fragmented sources of biomedical knowledge, including clinical studies, molecular biology databases, and pharmaceutical records. Manual analysis of these resources is time-consuming and often incomplete. To address this challenge, SMAG has developed a platform that integrates structured data into a unified, searchable framework.
Platform Overview
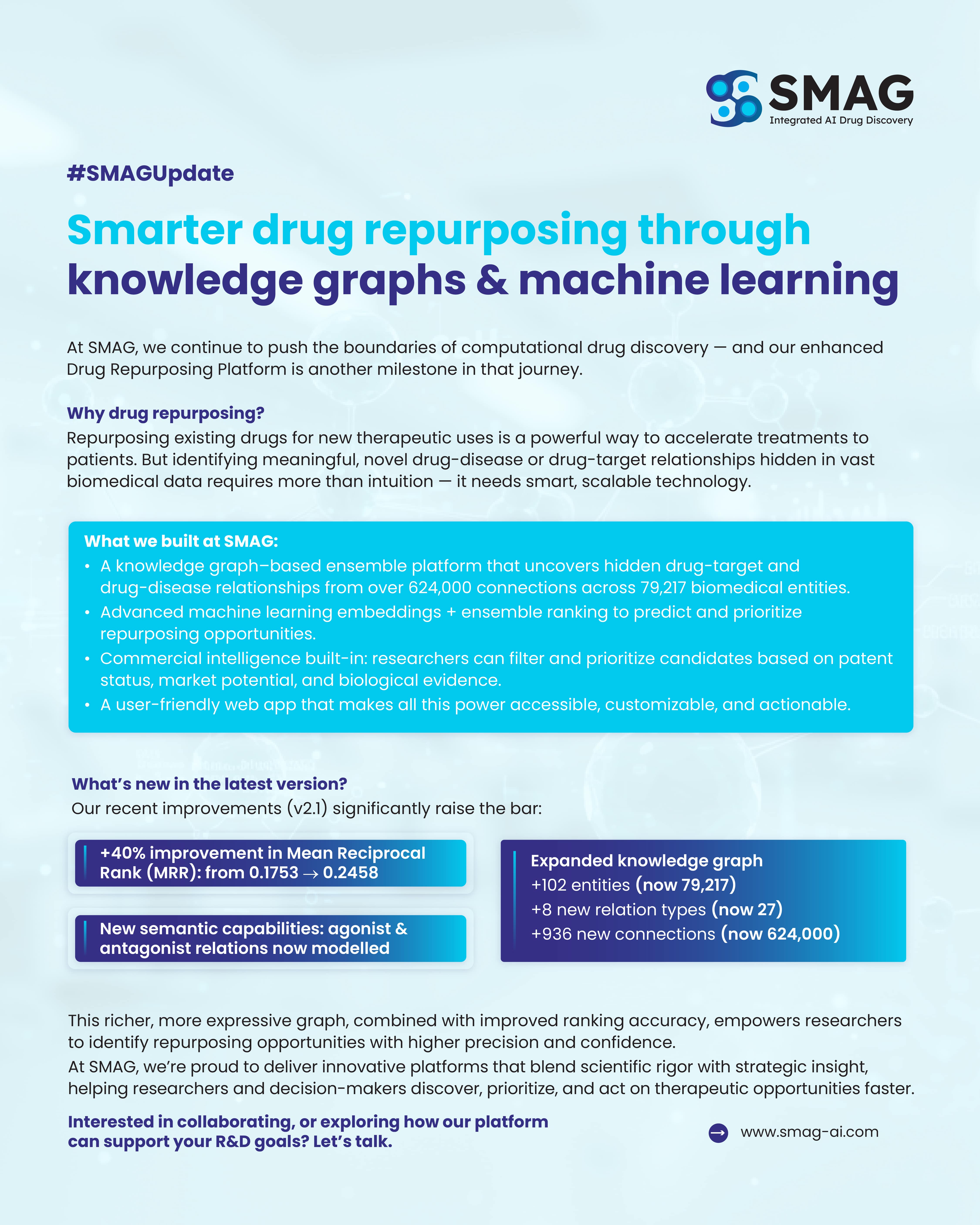
SMAG’s drug repurposing research is supported by a comprehensive biomedical knowledge graph that integrates data on drugs, diseases, targets, genes, and other key biological entities. This internally developed framework enables SMAG’s scientific teams to investigate potential drug-target and drug-disease relationships using evidence drawn from a wide range of curated biomedical sources.
Key Features:
1. Biomedical Knowledge Graph
- 79,217 entitiesacross various biological domains, including drugs, diseases, genes, pathways, and phenotypes
- 624,000+ connectionsmodeled through 27 relation types, such as "inhibits," "treats," and "interacts with"
- Newly introduced relation types include agonist and antagonist interactions, which provide greater pharmacological detail
2. Ranking of Repurposing Candidates
The platform includes a ranking system that prioritizes candidate drugs based on the strength and relevance of their relationships within the knowledge graph. This system evaluates multiple types of evidence, including biological plausibility, literature support, and connectivity within the graph.
In version 2.1, ranking performance has improved significantly, with a 40% increase in Mean Reciprocal Rank (MRR) — rising from 0.1753 to 0.2458. This metric reflects the improved quality of top-ranked results, allowing researchers to identify high-priority candidates more efficiently.
3. Commercial and Scientific Filtering Tools
Recognizing the importance of practical constraints in drug development, the platform allows users to filter repurposing candidates based on:
- Patent status
- Market potential
- Biological evidence scores
These filters help researchers and strategic teams align their selection with both scientific feasibility and development considerations.
4. Accessible Web-Based Interface
The web-based application provides a user-friendly environment for:
- Searching and filtering drugs and diseases
- Viewing connections and interactions in the knowledge graph
- Generating and exporting reports
- Tracking changes across different versions of the platform
What's New in Version 2.1
Version 2.1 includes several major updates:
Expanded Graph Structure
- +102 new entities added to the graph
- +8 new types of relationships
- +936 new connections, improving the resolution of drug-target and drug-disease networks
Improved Relationship Modeling
- Incorporation of agonist and antagonist relationships, improving the representation of pharmacological mechanisms
Enhanced Candidate Ranking
- Substantial improvement in ranking accuracy, with MRR performance increasing by over 40%
These improvements provide a richer foundation for hypothesis generation and prioritization in drug repurposing workflows.
Applications in Research and Development
The SMAG platform is suitable for use across multiple stages of pharmaceutical and biomedical research:
- Early-stage discovery: Identifying underexplored therapeutic uses for known compounds
- Preclinical evaluation: Prioritizing candidates based on biological relevance and development feasibility
- Clinical repositioning: Exploring indications with available safety data to support faster trial design
- Academic and institutional research: Generating research hypotheses supported by computational evidence
Conclusion
By integrating large-scale biomedical data into an accessible and structured platform, SMAG supports the systematic identification of new drug applications. The enhanced Drug Repurposing Platform (v2.1) delivers more accurate candidate ranking, expanded pharmacological modeling, and robust commercial filtering tools — all within a web-based environment designed for research professionals.
We invite researchers, healthcare developers, and scientific institutions to explore how this platform can support their work in discovering and validating new therapeutic pathways.
Contact Us
To learn more about SMAG’s Drug Repurposing Platform or to request a demonstration, please contact our team.