Accurate pharmacokinetic (PK) predictions are essential in modern drug discovery, where every decision impacts safety, efficacy, and development timelines. Among PK parameters, the Volume of Distribution at Steady State (VDss) is particularly important, as it reflects how a drug disperses throughout the body.
To accelerate this process, SMAG (SMart Analytics for Genomics & Drug Discovery) has developed a VDss Prediction Model, a next-generation computational tool built on ensemble learning. This model helps researchers predict VDss with high accuracy, offering a faster, cost-effective alternative to traditional laboratory methods.
Why VDss Prediction Matters
The Volume of Distribution (VDss) indicates how extensively a compound spreads beyond the plasma into body tissues.
- A high VDss suggests widespread tissue penetration.
- A low VDss indicates the compound largely remains in circulation.
This value is critical for:
- Drug dosing: Ensuring safe and effective therapeutic levels.
- Bioavailability modeling Predicting systemic exposure.
- Translational studies: Supporting extrapolation from preclinical to clinical settings.
Conventional experimental measurement involves intravenous dosing and plasma analysis, which can be time-consuming, resource-intensive, and impractical at early discovery stages. In silico models provide a scalable solution, enabling rapid compound screening.
Inside the SMAG VDss Prediction Model
The SMAG VDss model integrates cheminformatics and ensemble learning for robust, scalable predictions.
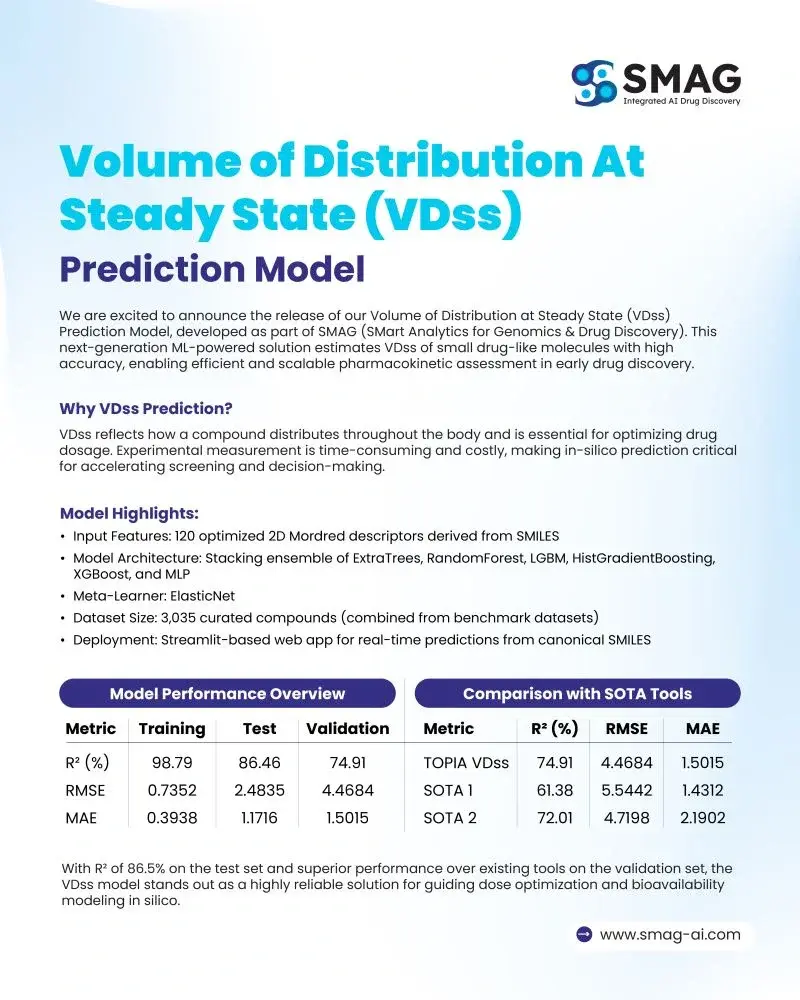
Model Highlights
- Input Features:120 optimized 2D Mordred descriptors derived from SMILES
- Base Algorithms:ExtraTrees, RandomForest, LightGBM, HistGradientBoosting, XGBoost, and MLP
- Meta-Learner:ElasticNet for refining ensemble predictions
- Dataset:3,035 curated compounds compiled from benchmark datasets
- Deployment:Streamlit-based web application enabling real-time predictions directly from canonical SMILES
This combination balances predictive performance, interpretability, and ease of use.
Model Performance
Rigorous benchmarking shows strong accuracy and reliability across training, test, and validation datasets.
Performance Metrics:R² (Coefficient of Determination), RMSE (Root Mean Squared Error), MAE (Mean Absolute Error)
| Metric | Training | Test | Validation |
|---|---|---|---|
| R² (%) | 98.79 | 86.46 | 74.91 |
| RMSE | 0.7352 | 2.4835 | 4.4684 |
| MAE | 0.3938 | 1.1716 | 1.5015 |
Comparison with SOTA Tools
| Model | R² (%) | RMSE | MAE |
|---|---|---|---|
| SMAG VDss | 74.91 | 4.4684 | 1.5015 |
| SOTA 1 | 61.38 | 5.5442 | 1.4312 |
| SOTA 2 | 72.01 | 4.7198 | 2.1902 |
Key Insights:
- High accuracy:R² = 86.46% on the test set.
- Better generalization: Outperforms state-of-the-art (SOTA) tools on validation.
- Lower error metrics:Reduced RMSE and MAE compared to benchmarks.
Applications in Drug Discovery
The SMAG VDss model can be applied across multiple stages of R&D:
- Early-stage screening - Estimate distribution profiles without experimental testing.
- Dose optimization - Support selection of appropriate therapeutic dose ranges.
- ADMET profiling - Integrate with absorption, metabolism, and toxicity models.
- Candidate prioritization - Focus resources on compounds with favorable distribution properties.
Looking Ahead
The VDss prediction model is a part of SMAG’s growing TOPIA Modeling Suite, which provides validated, computationally driven solutions for drug discovery. Our mission is to empower scientists with faster, data-backed decision-making tools that improve efficiency and reduce bottlenecks in early pharmacokinetics.
Frequently Asked Questions (FAQs)
Conclusion
The SMAG VDss Prediction Model represents a significant advancement in pharmacokinetic modeling. By delivering fast, accurate, and scalable VDss predictions, it reduces reliance on costly experiments and enables researchers to prioritize promising drug candidates with confidence.
Interested in learning more or collaborating? Contact us at [email protected].