
An update on SMAG’s machine learning-driven approach to early-stage pharmacokinetics
In our continuous effort to streamline and strengthen early drug discovery, SMAG has developed a Plasma Protein Binding (PPB) Prediction Model, designed to help researchers better understand drug distribution faster and with greater efficiency.
This development reflects our broader mission at SMAG: to translate complex biomedical challenges into practical, computational solutions that improve research workflows and decision-making in pharmaceutical R&D.
Why Plasma Protein Binding Matters
Plasma protein binding is a critical pharmacokinetic property that describes how much of a drug binds to proteins (like albumin) in the bloodstream. Only the unbound (free) portion of a drug is pharmacologically active, so PPB directly influences:
- Drug distribution
- Clearance rate
- Therapeutic efficacy and safety
Traditional methods for measuring PPB rely on experimental procedures such as equilibrium dialysis or ultrafiltration — which, although accurate, are time-consuming, labor-intensive, and often not scalable during early-stage screening.
SMAG’s Approach to PPB Prediction
To support faster and broader evaluation during preclinical research, our team developed a machine learning-based PPB prediction model that combines accuracy with usability.
Key Features:
- ML-Driven Ensemble Model:
Built using an ensemble of advanced machine learning algorithms optimized for regression tasks.
- Diverse Training Data
The model was trained and validated on a wide-ranging collection of publicly available datasets, ensuring broad chemical diversity and generalizability.
- Simple Input, Real-Time Output:
Researchers can input a compound's SMILES string (a text representation of its molecular structure) and instantly receive predicted PPB values streamlining the screening of new compounds.
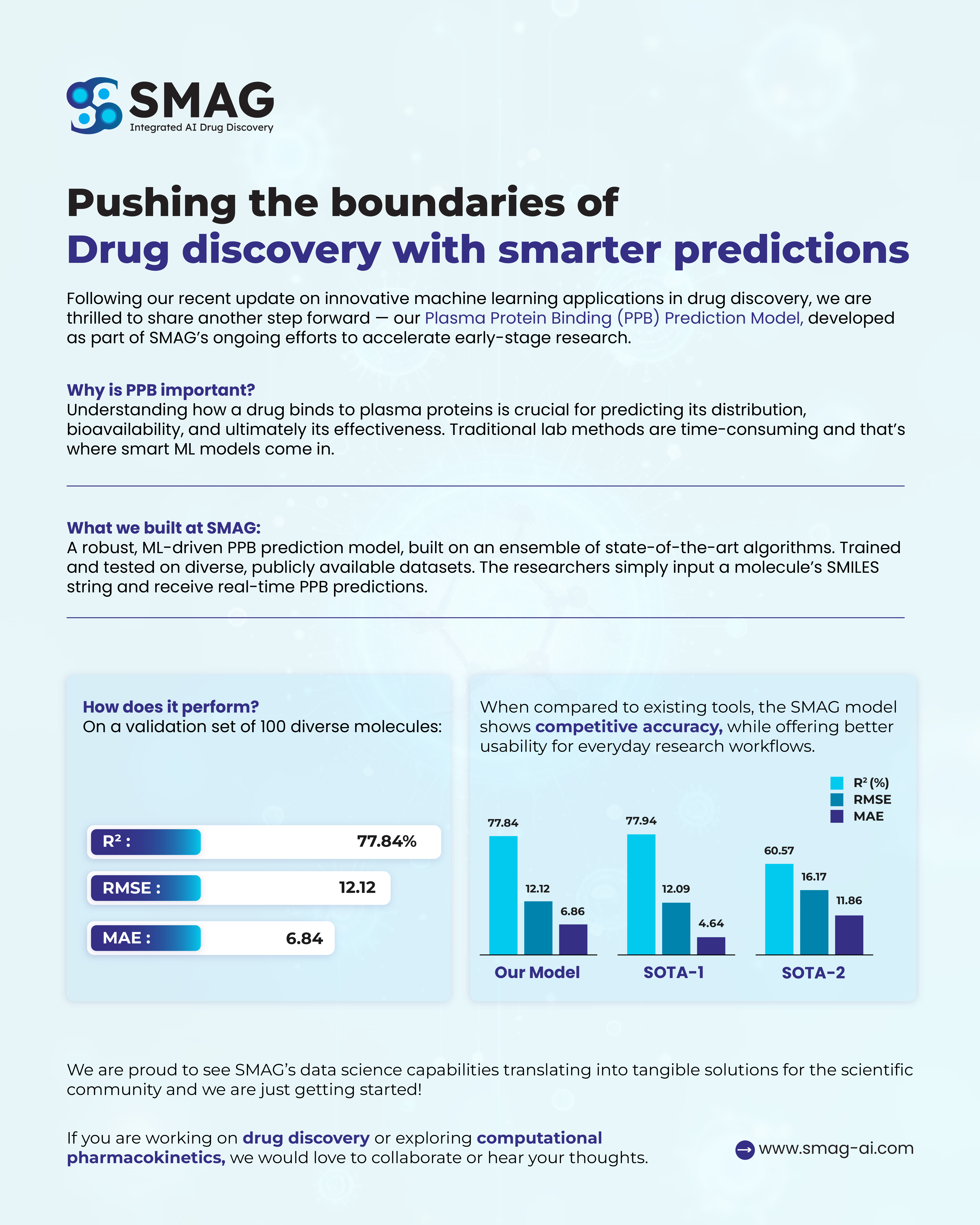
Performance Overview
Our internal benchmarking shows that SMAG's PPB prediction model performs competitively against leading state-of-the-art (SOTA) tools, with robust statistical results across key evaluation metrics.
| Model | R² (%) | RMSE | MAE |
|---|---|---|---|
| SMAG Model | 77.84 | 12.12 | 6.84 |
| SOTA-1 | 77.94 | 12.09 | 4.64 |
| SOTA-2 | 60.57 | 16.17 | 11.86 |
- R² (coefficient of determination): Measures how well predicted values match observed outcomes
- RMSE (Root Mean Square Error):Captures average error magnitude
- MAE (Mean Absolute Error): Indicates average prediction error
While SOTA-1 offers slightly higher precision in MAE, the SMAG model delivers comparable accuracy with significantly improved ease of use and integration into day-to-day research workflows.
Built for Real-World Research
Beyond its technical performance, the strength of the SMAG PPB model lies in its accessibility and speed, offering practical value for medicinal chemists, computational biologists, and pharmacokinetics researchers.
Use cases include:
- Early screening of candidate molecules
- Predicting PPB during lead optimisation
- Complementing in vitro testing with computational insights
The tool is intended to support internal discovery teams while also paving the way for future collaborative research in computational pharmacokinetics.
Looking Ahead
At SMAG, we’re proud to see our in-house data science and pharmacology efforts translate into working tools that accelerate decision-making and reduce bottlenecks in early drug development.
This PPB prediction model is one of several initiatives underway as we continue to build integrated, intelligent systems for drug discovery.
Interested in Collaborating?
If your team is working in drug discovery, pharmacokinetics, or computational modelling, we’d love to connect. Whether it’s exploring joint research opportunities or sharing feedback on our approach, collaboration is key to advancing meaningful progress in this space. Contact Us Now